Ringmaster Of Craniofacial Development- Neural Crest Cell
A Bansal, R Bansal, D Shetty, A Manchanda, P Aggarwal
Keywords
ectomesenchyme, morphogenesis, neural crest cells
Citation
A Bansal, R Bansal, D Shetty, A Manchanda, P Aggarwal. Ringmaster Of Craniofacial Development- Neural Crest Cell. The Internet Journal of Dental Science. 2009 Volume 9 Number 2.
Abstract
Embryonic development depends on the precise temporal and spatial interactions between various cells and extracellular molecules. One of the most fascinating characteristics of embryonic development is the migratory and differentiative potential of neural crest cells (NCC). The current article reviews the interactions that occur between extracellular matrix components and cranial neural crest cells during the differentiation of craniofacial skeletal elements and its role in tooth morphogenesis and its patterning.
Introduction
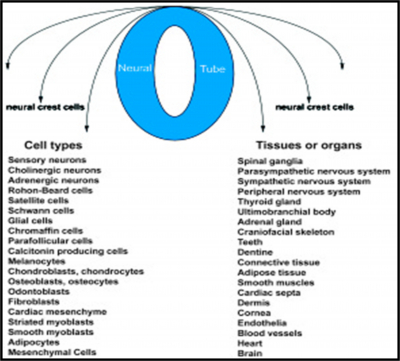
Embryonic development depends on the precise temporal and spatial interactions between various cells and extracellular molecules. One of the most fascinating characteristics of embryonic development is the migratory and differentiative potential of (NCC).1-3 Neural crest cells from the roof plate of the neural tube undergo an epithelial to mesenchymal transition, delaminating from the neuroepithelium and migrating through the periphery where they differentiate into varied cell types (Figure 1).4 In the trunk, these include the peripheral nervous system, melanocytes, and glandular structures. In the craniofacial region, in addition to forming the aforementioned tissues, the neural crest cells are also the major source of the mesenchymal population that differentiates into osteoblasts, chondroblasts, and odontoblasts. This additional skeletogenic potential of the cranial neural crest makes it qualitatively different from trunk neural crest and, in some ways, similar to mesodermal tissue.
Figure 1
NCC were identified first in 1868 in the chick embryo by Wilhelm His, who designated it as Zwischenstrang, literally an ‘intermediate cord’.5, 6 Although it was initially associated with the origins of neurons and ganglia, Julia Platt demonstrated in the 1890s that the visceral cartilages of the head and dentine forming cells of the teeth in the mud puppy Necturus, also arise from the neural crest.7
The current article reviews the interactions that occur between extracellular matrix components and cranial neural crest cells during the differentiation of craniofacial skeletal elements and its role in tooth morphogenesis and its patterning.
Branchial Arch formation: Oral Ectoderm and Ectomesenchyme
The accumulation and proliferation of cranial neural crest (CNC) cells, together with proliferation of the ectoderm cells, produce a series of swellings that constitute the facial primordia. Mammalian teeth develop on two of these primordia: the front nasal process and the first branchial (pharyngeal) arch, which forms the mandible and proximal maxilla. These primordial structures form as CNC cells from the developing brain and accumulate beneath the ectoderm, covering what will become the face and oral cavity.8 The lineage origins of oral ectoderm cells in mammals have not been accurately traced, but the anterior neural ridge rostroventral to the migrating CNC cells yields the neural epithelium of the head, including the olfactory placodes, Rathke's pouch, and the oral epithelium.9,10 Teeth develop from the rostra (oral) cells of the developing mandibular primordium, whereas bone and cartilage (Meckel's) develop more caudally (aboral). The different positional fates of these mesenchymal cells are determined early in the formation of the primordium. Recombination experiments have confirmed that oral ectoderm is the source of inductive signals for tooth development and for establishing odontogenic competence in oral ectomesenchyme.11 Expression of the closely related LIM-domain homeobox genes Lhx6 and Lhx7 is restricted to oral ectomesenchyme of the mandibular and maxillary processes and complements that of Goosecoid (Gsc), which is expressed in aboral mesenchyme.12 The ectoderm appears to be involved in the induction of both oral and aboral mesenchymal gene expression.
The ectoderm expresses a wide range of signalling molecules, fibroblast growth factors (Fgfs), bone morphogenetic proteins (Bmps), Wnts, and hedgehog proteins (HHs), and it is the restriction of Fgf8 expression to the oral ectoderm that appears to establish the antero-posterior axis of the first branchial arch.13, 14 The restriction of Gsc expression to aboral mesenchyme involves repression by Lhx6/7-expressing cells, although the mechanism that restricts Lhx6/7 expression to oral mesenchyme is independent of Gsc and is more probably related to the distance from the source of FGF8. Targeted mutations in Lhx6 or Lhx7 do not result in any tooth defects; such defects may be revealed only when these mutations are combined.15 Mutations in Gsc, however, do have a profound mandibular bone phenotype with severe truncation, but the teeth develop normally. Endothelin 1 is expressed in the entire mandibular epithelium and appears to act as a maintenance factor for Gsc expression. Both endothelin 1 and endothelin receptor knock-outs have a mandiblular phenotype similar to that of Gsc.16
The ectomesenchyme cells of the developing facial processes that participate in tooth development form from CNC cells. Lineage and cell fate analysis has demonstrated a great array of cellular fates arising from CNC cells, including neurons, neuroglia, smooth-muscle cells, calcitonin-producing C-cells, melanocytes, adipocytes, tendon connective tissue cells, mesenchymal cells, fibroblasts, cementoblasts, odontoblasts, chondroblasts, chondrocytes, secondary chondroblasts (in secondary cartilage), osteoblasts, and osteocytes.17, 18 The unique ability of cranial neural crest cells to develop into hard skeletal tissue distinguishes them from trunk neural crest cells in higher vertebrates, whose cartilage and bone elsewhere in the body have a mesodermal origin.
Determination of tooth morphogenesis
Mammalian tooth morphogenesis must be controlled by either ectodermally derived and/or ectomesenchymal derived cells, since these are the only cell types that form teeth. Evidence suggests that the information for generation of an incisor or a molar form tooth is inherent in the ectomesenchyme, whereas the establishment of this information and the actual mechanics of employing this information are carried out by ectodermally derived cells of the enamel knot.8 During the initiation of odontogenesis, localized signaling is important for growth and development of the tooth bud, whilst later during morphogenesis, Sonic hedgehog plays a role in cellular differentiation and polarization in the epithelial component of the tooth germ.19
The odontogenic homeobox code
The idea that the shape of each tooth is specified by a code of different home proteins was proposed from observations of the region-specific, overlapping domains of several homeobox genes in ectomesenchyme.20 (Figure 2)21 In particular, expression of Dlx-family members and Barx l is restricted to proximal (presumptive molariform) ectomesenchyme, whereas expression of genes such as Msx l and Alx 3 is absent from proximal regions and is restricted to distal (presumptive incisor) ectomesenchyme.22 Stated simply, the combination (code) of homeobox genes expressed in cells determines the morphogenetic pathway a tooth germ will follow. Thus, cells expressing Dlx and Barx l genes (but not Msx l or Alx 3) are directed to follow a molar pathway of morphogenesis, whereas cells expressing Msx l and Alx 3 but not Dlx or Barx l will be directed to follow an incisor pathway.
Figure 2
Three important conclusions were derived from this model. The first is that there is not one specific gene for each tooth type. Second, the code is both positive and negative; thus, the absence of a gene is as important as its presence. Third, the code is overlapping and can thus provide morphogenetic cues for many different tooth shapes.
Teeth develop by a reciprocal series of epithelial-ectomesenchymal interactions that have been identified by recombination and bead implantation experiments.23 Spatial domains of homeobox genes are established in the ectomesenchyme and provide the positional information (odontogenic homeobox code) for the specification of tooth shape. Two possible mechanisms for the regulation of these restricted domains of ectomesenchymal gene have been put forth (1) neural crest cells contain a pre-pattern; and (2) neural crest cells respond to positional signals from the oral epithelium. Removal of epithelium from mandibular arches at E10 or before, results in a total and rapid loss of almost all ectomesenchymal derived homeobox gene expression. The addition of FGF8 beads to the ectomesenchyme induces homeobox gene expression only around the bead, indicating not only that ectomesenchymal gene expression is dependent on epithelial signals but also that ectomesenchymal cells are plastic in their responses to FGF8 at this time, since cells not normally responding to FGF8 have the ability to do so. Removal of epithelium at E10.5 also results in loss of gene expression, but significantly, subsequent addition of FGF8 beads restores expression in the original expression domain only. Removal of epithelium at Ell does not affect gene expression, indicating that the spatial homeobox gene expression domains are established and maintained in the absence of epithelial signals. These results indicate that the signals that induce ectomesenchymal homeobox gene expression come from the epithelium and that, within the space of 24 hours, there is a dramatic change in how ectomesenchymal cells respond to these signals. Up to E10, all ectomesenchyme cells appear to be uncommitted and competent to respond to epithelial signals regardless of position. By E10.5, the spatial expression domains have been established in the ectomesenchyme by the action of epithelial signals, such as FGF8 and BMP4, and although these signals are still required to maintain these expression domains, the ectomesenchymal cells have lost their competence to express any gene in response to the signals. The spatial expression domains established at E10 are fixed and now maintained by the epithelial signals. By Ell, expression of ectomesenchymal genes does not require epithelial signals (Figure 3).8 Epithelial signals thus regulate the spatial expression of homeobox genes in the ectomesenchyme which in turn control morphogenetic pathways, probably by influencing enamel knot function. The control of tooth shape thus mirrors the general control of tooth formation, with information being passed from epithelium to ectomesenchyme and back again to epithelium.8
Role of Dlx in craniofacial development
Dlx genes in particular seem important for the morphogenesis of proximal jaw hard tissues and most specifically distinguish the upper from the lower jaw structures. Dlx5 and Dlx6 are co-expressed in proximal ectomesenchyme of the mandibular primordia in domains similar to Dlx l and Dlx 2. Significantly, however, Dlx 5 and Dlx 6 are not expressed in the maxillary arch.24 Mutations in Dlx l and/or Dlx 2 affect maxilla development, presumably because Dlx 5 and Dlx 6 genes carry out this function in the mandible in the absence of Dlx l and Dlx 2. The fact that these genes are differentially expressed in the mandible and maxillary arches indicates a basic positional difference between the ectomesenchymal cells.
Figure 3
A basic difference between ectomesenchyme cells of the mandibular and maxillary primordia was confirmed by cell transplantation experiments where expression of the Dlx genes was used as markers. Thus, transplants of maxillary arch cells (Dlxl/1 -positive; D/x5/6-negative) into the mandibular arch (DM/2-positive; D/x5/6-positive) results in the donor cells retaining their gene expression patterning, i.e., they do not start to express Dlx5/6. The reverse transplant of mandibular cells into the maxilla also resulted in the donor cells retaining their characteristic gene expression, i.e., they continued to express Dlx5/6 in an environment where they are surrounded by non- D/x5/6-expressing cells.25 Since the signal for expression of all four of these Dlx genes is FGF8, and this is present in the epithelium of both the maxillary and mandibular primordia, it suggests that the ectomesenchymal cells themselves respond differently. This suggests that the cranial neural crest cells that populate these two primordia are different from each other, but within each individual population, all the cells are uncommitted with respect to hard tissue morphogenesis.8, 26
Factors that mediate neural crest differentiation
Intrinsic to any discussion of the factors that mediate the differentiation of cranial neural crest cells into skeletal elements is determination of whether the neural crest cells are differentially committed to a particular lineage prior to, during, or after migration. This topic has been vigorously investigated for many years and is still controversial. Recently, vital dyes have been used to follow the migration of individual neural crest cells and have shown that the offspring of at least some cells has the capability of differentiating into more than one cell type suggesting that neural crest cell have pluripotential capability.27 Previous in vitro studies have suggested that the formation of cartilage and bone from neural crest derived ectomesenchyme is dependent upon prior interaction with embryonic epithelium. When this epithelium is enzymatically removed, the migrating neural crest cells do not differentiate into skeletal components. Epithelia from other embryonic sources can be substituted for the mandibular, but the substituting epithelium has inductive capabilities only during particular periods of embryogenesis, presumably during time periods when this epithelium has inductive capabilities for other systems. This reinforces the concept that the inductive agent for the formation of mandibular bone is dependent upon interaction with the mandibular epithelium. Tenascin/cytotactin is an extra cellular protein that has been localized to the migratory pathway of the neural crest and has been shown to be associated with chondrogenic and osteogenic tissues undergoing differentiation. It is present in restricted distribution in tissues whose differentiative fate has been determined, but not yet expressed phenotypically. It apparently interacts with fibronectin, resulting in cell rounding and condensation.28
Neural crest cells have been called the ‘explorers of the embryos’ because they are able to migrate extremely long distance, following specific pathways and colonizing almost all the tissues of the embryo. It has been found in recent years that many of the factors that belong to the NC genetic cascade are also involved in controlling tumour progression. Furthermore, cellular and embryological evidence seems to indicate that cancer cells share many characteristics with NC cells. The epithelial–mesenchymal transition process that both kinds of cells undergo is controlled by the same molecular machinery, including cadherins, connexins, Snail and Twist genes and matrix metalloproteases.28, 29
Neural Crest Cell Migration-Cell Tracing Techniques
Cell Marking Techniques: Early studies of embryonic cell lineage were based on natural markers found in certain cells, such as pigment granules, distinctive nuclear morphologies, or yolk inclusions, which could be used to trace the migration of and differentiation of cells. But this approach didn't prove useful as crest cells have got no distinctive natural markers. In 1963 Weston and 1967 Chibon devised a more precise technique by labelling the DNA of migrating neural crest cells with tritiated thymidine. Migration of cells labelled with tritiated thymidine can be followed by autoradiography (Autoradiography is used for visualizing a newly synthesized cell product after cells are exposed to radioactive precursor molecules for varying lengths of time either under in vitro or in vivo condition). However because the radioactive label is diluted each time a cell divides & synthesizes new DNA, the technique can trace a cell lineage through only a limited number of generations. So this technique is not stable and furthermore reuptake of label released by dead cells compromised specificity. The problem of label dilution was solved by development of quail chick chimera system.
Quail-Chick Chimera Technique: The technique is based on the observation by Le Douarin (1969) on the structure of the interphase nucleus in the Japanese quail. The cells of this species are characterised by the condensation of constitutive heterochromatin into a single mass, generally centro nuclear, associated with the nucleolus. Such a characteristic is rare in animal kingdom. In most species the constitutive heterochromatin is evenly distributed in the nucleoplasm in small chromocenters e.g. in chick. Quail and chick cells can thus easily be distinguished by DNA staining or by electron microscopy, where the large DNA rich nucleolus of the quail is easily recognisable. These inter-species differences have been used to study the migration of embryonic cells and to analyze the contribution of cells of different embryonic origins to complex tissues or organs during ontogeny. Since the quail and the chick are closely related in taxonomy, the substitution of definite territories between embryos of these two species in ovum results in viable chimeras which develop normally and can hatch. Thus when definite fragments of either the neural fold or the neural tube and associated neural crest of the quail are grafted isotopically or is chronically into the chick (or vice versa) the migration and fate of their constituted cells can be followed. This approach has been systematically applied to whole neuraxis to establish the fate map. By grafting cranial paraxial mesenchyme and the 6 rostral somites Couly, Coltey and Le Douarin (1992), were able to delineate precisely the respective contributions of the somites, paraxial mesenchyme and the neural crest to skull. The question whether crest cells are, unipotent, pluripotent or totipotnet has been partly answered by the experiments in which quail cell from one part of the neural crest were transplanted to various locations along the chick neural tube. The interpretive problems of thymidine and quail-chick marking experiments can be avoided by studying single cells marked with fluorescent dyes.
Cell Lineage Study Using Fluorescent Dye: It is now possible to follow the development of a single neural crest cell by injecting it with a highly fluorescent, non-toxic marker that remains visible in the injected cell and its descendents. In some cases, a single neural crest cell injected with lysinated rhodamine dextron (LRD), before or during migration gives rise to daughter cells that develop into a variety of neural crest derivatives. Clonal analysis of LRD-injected trunk neural crest cell precursors of the mouse shows that their developmental potential depends, in part, on the time that they emigrate from the neural tube. Single cells injected during early neural crest migrations may give rise to neural tube cells as well as neural crest derivatives that migrate along both dorsal pathways and ventral pathways. Derivatives of the dorsal pathway include melanocytes and cells of the dorsal root ganglia, while cells of the ventral pathway form sympathetic ganglia and adrenomedullary cells. In contrast migration of late emigrating neural crest cells is restricted to dorsal pathway (Larsen; 1997).
Cell Lineage Studies Using Retroviruses: In amphibians and avian, direct cell injection of fluorescent lineage tracers is possible. This is so far impossible in mammals owing to inaccessibility of the embryos in utero. Instead, lineage analysis has relied on the injection of progenitor cells in the ventricular zone on rodent embryos using replication defective retroviruses (Sanes, Rubinstein and Nicolas, 1986). The retroviral DNA incorporates into the genome of the infected cell providing an indelible marker that is transmitted to all progeny during cell division, but not to surrounding cells. Retroviral techniques have yielded some information about lineage in the various regions of the nervous system. Very recently techniques using replication defective virus have been used for developing heart. In this study a replication defective avian spleen necrosis virus which lacks the viral structural genes gag, pol and env, (which have been replaced with the bacterial B-galactosidase gene lac Z), has been used to mark NC cells. Its results were compared with those of chicken-quail chimera used as a gold standard. Retrovirus infected cells expressed the Lac Z gene and could be distinguished both in whole mount preparation and in the histological sections. Because of the possibility of whole mount preparations retroviral approach using B-gal for staining is far superior to the chick-quail chimera to obtain overall picture of labelled cell progenitor in the target organs
Transgenic Techniques: Recombinant DNA techniques can now be used to study differentiation in specific tissue lineages. In an experiment a bacterial transgene was introduced into the mouse genome in such a way that it caused certain cell of neural crest origin - specifically neurons of dorsal root ganglia to stain themselves blue. The lac Z gene was inserted into the transgene in a position that put it under the control of the same regulatory DNA region that controls the expression of the gene for a neurofilament protein called peripherin. Copies of this engineered sequence were then injected into the male pronucleus of pronuclear stage mouse embryos, where they become incorporated into the embryonic genome. Peripherin is normally expressed in a number of tissues of neural crest origin. The lac Z gene in the transgenic mice should be activated in all cells that express the endogenous peripherin gene, with the result that all peripherin producing tissue would stain blue.
Marker Technique: Recently a number of markers have become available that label migratory neural crest cells. These include antibodies or probes to the receptor tyrosine kinase - Ret(Pachnis, Manko and Constantini; 1993, Tsuzuki., et al 1995., Dubec et al 1996., Wantanabe et al 1997) the low affinity neurotrophin receptor, P 75 NTR (Baetge & Gershon; 1990; Chalazonitis, et al 1994; Lo and Anderson, 1995), the endothelin-B receptor (Gariepy et al 1998) the 5-Ht2 B receptor (Florica- Howells et al 1998) the transgene DBH n lac z (Kapur, et al 1992) the catecholamine synthetic enzyme, tyrosine hydroxylase TH (Cochard, et al 1978; Teitelman et al 1978; Jonakait et al 1979) and the transcription factors MASH1 (Lo, et al 1991; Lo and Anderson, 1995; Lo, et al 1997), Phox 2a(Tiveron, et al 1996), phox 2b (Pattyn et al 1997), and sox 10 (Herbaerth, et al 1998). All the abovementioned antibodies or probes are concerned with enteric nervous system.
The markers or probes for melanocytes are MEBL-1 (Kitamura et al 1992), TRP-1 (Reddy, Faraco & Erickson; 1998), TRP-2 (Steel, et al 1992) Mitf (Opdecam et al 1997) and C-Kit (Kitamura et al., 1992; Wehrle-Haller and Weston., 1996.) For Neural crest derived neurons the markers are Hu (Marusich and Weston., 1992; Marusich, et al 1994), SC-1/BEN (Pourguie, et al 1990), and Neuron specific Tubilin (Lee, et al 1990). For Neural crest derived Glial cells-7B3 (Henion, et al 2000) and for Trunkal neural crest cells – NC1/HNK1 (Tucker et al 1984) are the markers.
Conclusion
Here, we have concentrated on the interactions that occur between extracellular matrix components and cranial neural crest cells during the differentiation of craniofacial skeletal elements. Cell-cell signalling pathways and their target nuclear factors have been identified as key mediators of the progressively complex exchange of information between ectoderm and ectomesenchyme. The constantly changing direction of the reciprocal signalling and cell responses between ectoderm and ectomesenchyme enables cells to monitor their relative spatial positions and differentiated states continuously. The dramatic effects of tinkering with gene regulation on body form have revealed many of the mechanisms that regulate morphological evolution and the challenge in the future will be to visualize the genetic and cellular interactions occurring in the neural crest cells of rapidly evolving populations, which should aid our understanding of the specific factors regulating selection driven evolution.